A tool for modeling biological systems
Bioframer combines novel user interface paradigms with new foundations for computer reasoning, to model information at the level of context.
A key aspect of this work is sponsored by the National Academies Keck Futures Initiative (NAKFI): we visualize systems as systems, as well as their interactions and components.
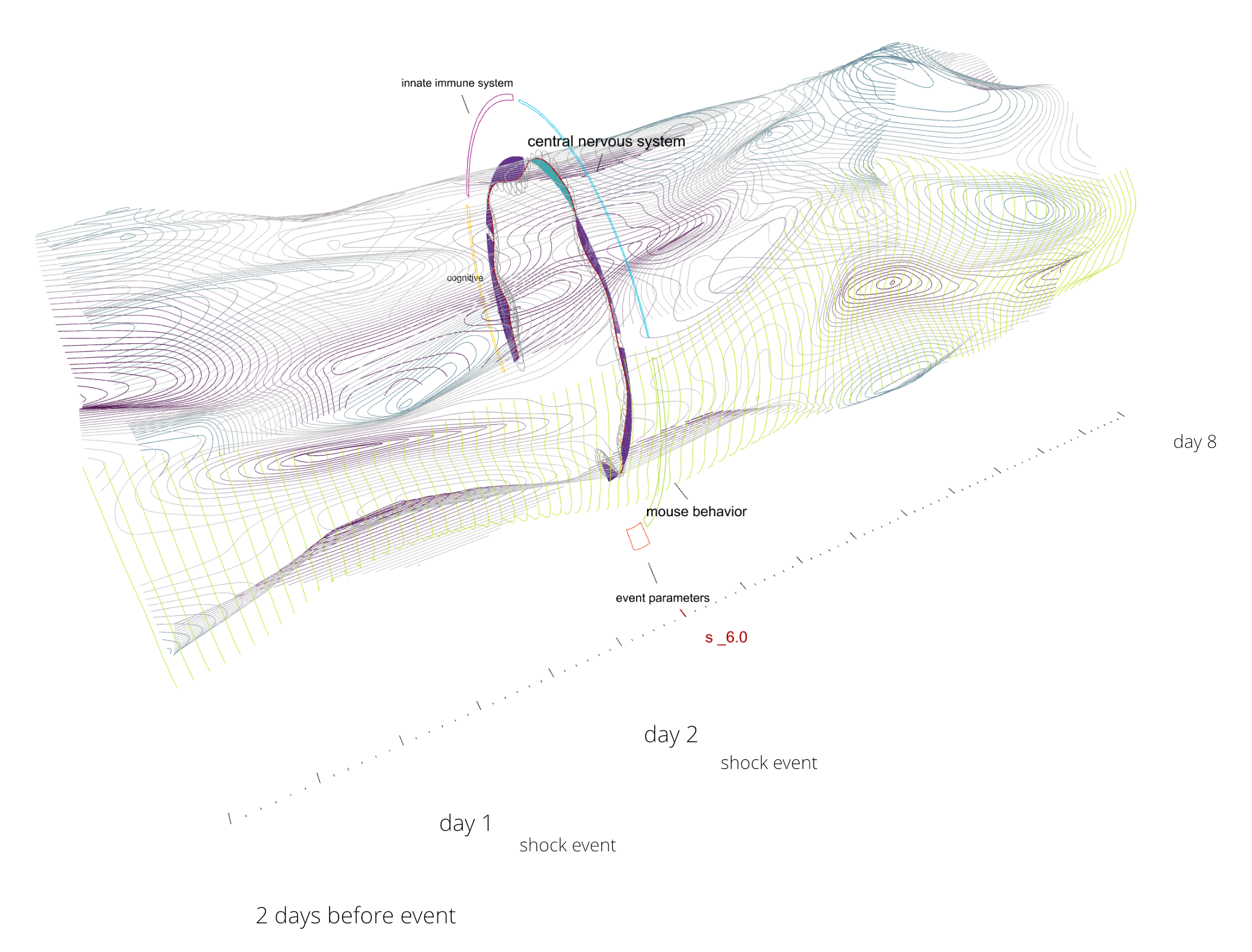
One target scenario: In post-traumatic stress disorder (PTSD), cognitive and biophysical systems interact to produce the symptoms of stress and sleep loss. Resolving these symptoms requires an understanding of how cognitive and biophysical systems govern each other when unbalanced. These shifts will be visualized as streams of information which interact and flow. Forms of information that currently elude bioinformatic modeling will be included: unknowns, ambiguity and retroactive reinterpretation. It will also be possible to zoom into the behavior of these communal systems, to discover when individual actors like neurons and their signal neurotransmitters are acting as causal agents across multiple processes.
Modeling interactions at the level of context is possible due to the novel combination of three approaches:
Situation Theory enables deeper causal modeling than current tools allow. It is based on advances in mathematical logic to handle contextual reasoning.
Geometric Logic supports practical coding to support integration with legacy systems, cloud processing and big data open source resources.
Design in Information Flow provides new user interface paradigms, addressing the problem of how to expose and manipulate the interactions between multiple contexts or systems.
Our overall goal is to empower small biomedical teams to enable a greater understanding of causal triggers among levels and systems, making it possible to plan more targeted experiments.
Collaborators are:
– Beth Cardier (knowledge modeling), Sirius-Beta Corp.
– Niccolo Casas (architecture and design), RISD Rhode Island School of Design; University College of London.
– Alessio Erioli (engineering) Università di Bologna.
– Ted Goranson (system design), Sirius-Beta Corp.
– Patric Lundberg (biomedical virus research), Eastern Virginia Medical School.
– Larry Sanford, (biomedical sleep and trauma research), Eastern Virginia Medical School.
– Richard Ciavarra, (biomedical virus and cancer research), Eastern Virginia Medical School.
_____
In Bioframer, we envision two main types of views. The most traditional presents facts arranged from left to right, with links that denote what ’causes’ what. Some novelties here are that nodes can be nested/exploded for examination; nodes can contain unknowns or partial knowns; any element’s formal definitions can be examined and edited. When doing so, causal relationships may change.
The ability to model retroactive cause is novel in itself. What’s radical is that the shape of this 3D object is influenced by situation governance. ‘Pulling’ by the user can explore and change internal tensions.
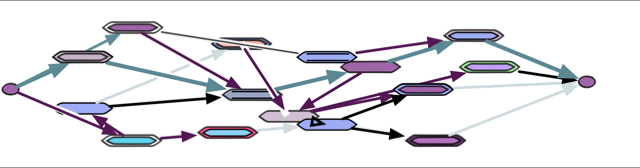
A high level causal lattice view is:
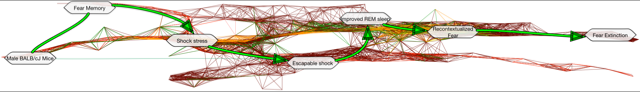
An exploded lattice can be quite complex. The primary path is imaged above:
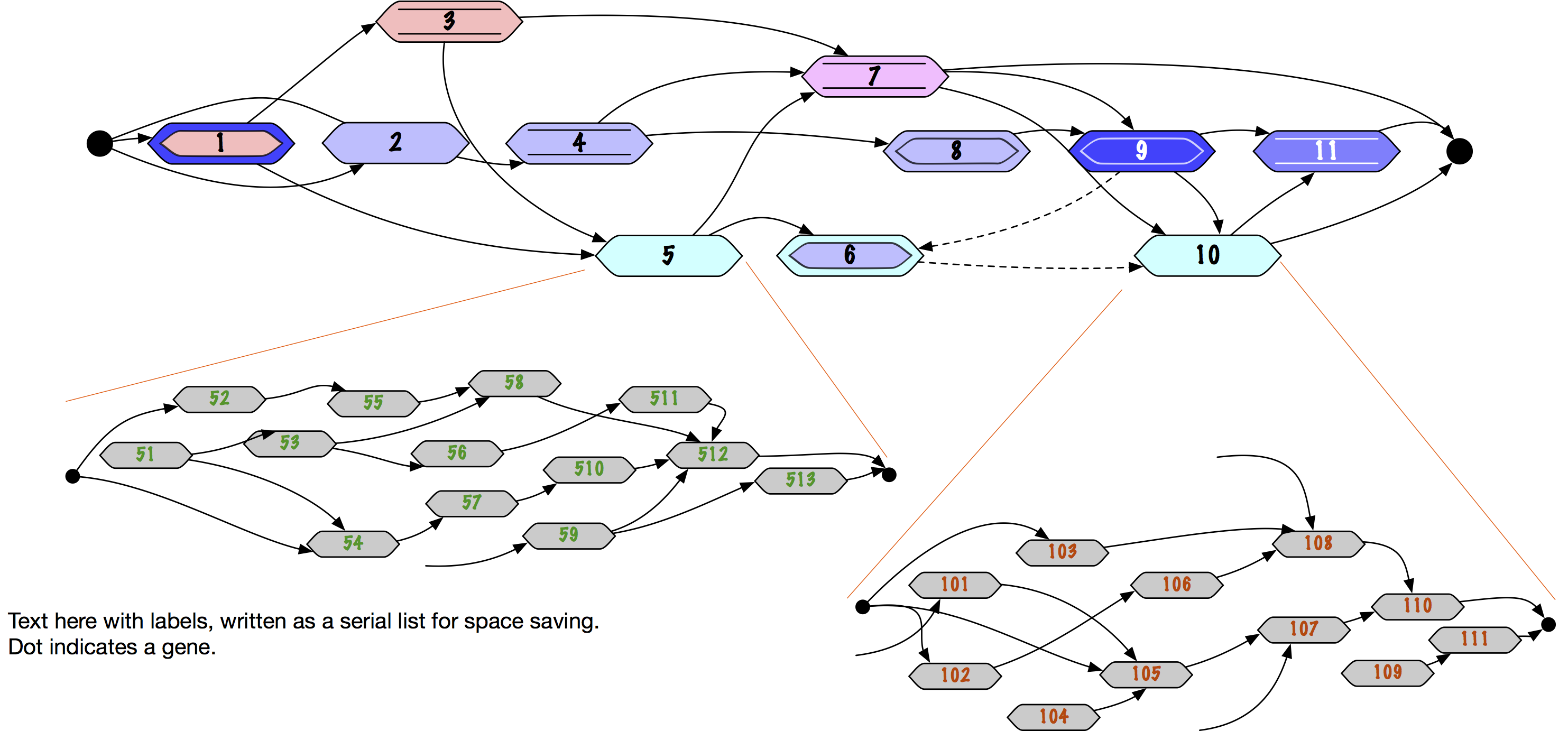
It’ll be possible to zoom between levels:
The other view is the subject of the NAKFI work, and is more challenging. How do we display the ‘clouds’ of situations themselves and their influence on the lattice? Below are some studies for a shell to surround the lattice. It too will be a 3D navigable object. Studies for this shell are shown below.
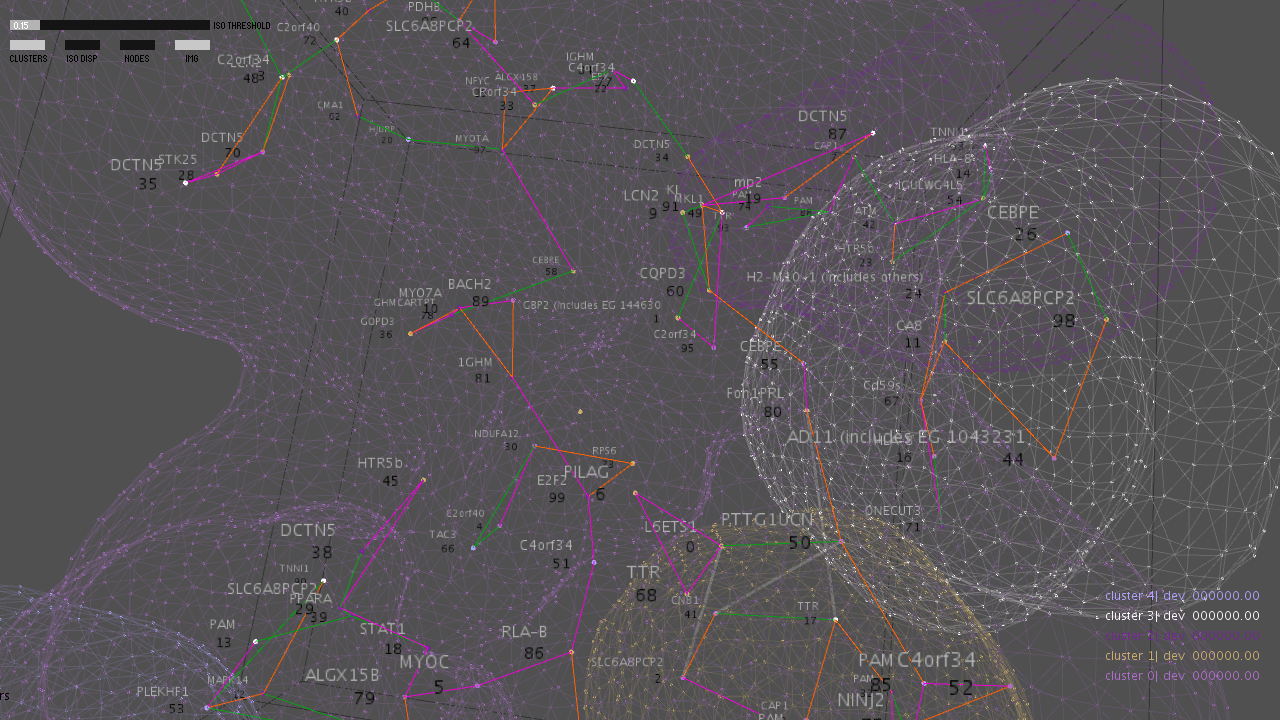
A study for the visualization.
_____
Some papers on progress toward this tool, with explanations of the figures are:
More information about the National Academies Keck Futures Initiative can be found on the NAKFI website and Beth’s blog.