Building a Narrative Modeling Tool:
Capturing Implicit Contextual Influence

This project builds a narrative modeling platform that can capture semantic information and its inferences as they change. One goal is to capture a new tier of information that usually can’t be included in models, such as social, emotional and learning information. Another is to model connections between diverse disciplines, such as the psychological, neurological and microbiological information entailed in understanding how stress impacts on the immune system. A third is to newly map how information is structured at the level of systems: how new ideas emerge, evolve and interact.
The modeling platform is being built in Unity to leverage 3D so that new information architectures are able to be built and explored.
This is a joint initiative between Sirius-Beta and the Virginia Modeling Analytics and Simulation Center, Old Dominion University. It is based on the design foundations developed during the National Academies Keck Futures Initiative project, Design in Information Flow (described below).
Updates on this project can be found on my blog, here.
A short overview can be found here.
Collaborators:
Beth Cardier, Sirius-Beta Corp.
Ted Goranson, Griffith University, Australia.
Saikou Diallo, John Shull, and Alex Nielsen, Virginia Modeling Analytics and Simulation Center, Old Dominion University.
Niccolo Casas, RISD Rhode Island School of Design; The Barlett College, University College of London
Patric Lundberg, Richard Ciavarra and Larry Sanford, Eastern Virginia Medical School.
These next two projects, both funded by the National Academies Keck Futures Initiative, solve problems in different domains: computational reasoning (Design in Information Flow) and biomedicine (Towards the Innernet).
Design in Information Flow:
Using aesthetic principles to overcome computational barriers in the analysis of complex systems.
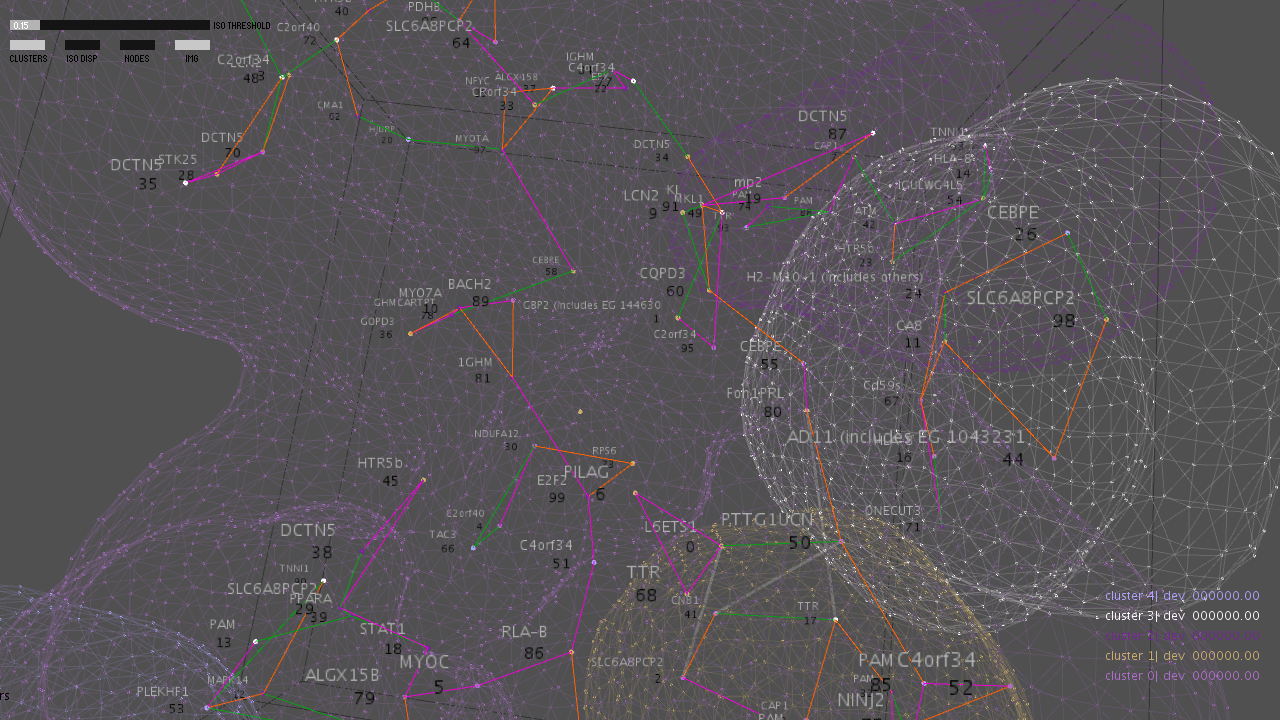
This project models streams of biomedical information as evolving form. We discover new methods of contextual integration using artistic mechanisms to match and constrain the available information: analogical structure, optimal tension resolution and the aesthetics of design. These operations are supported by the philosophy and new formalisms for situation theory, based at Stanford.
The goal is to overcome the barriers in computational logic that would usually prevent different kinds of data from interacting.
This work is supported by a $50K seed grant from the National Academies Keck Futures Initiative. It will occupy new territory somewhere between art, biomedicine and informatics. A long-term target is to develop a new visualization tool for small biomedical teams.
Details about this project and our general approach can be found here.
The final report for the project can be found here.
A gallery of images produced by the project can be found here.
I discuss interdisciplinary techniques learned during this project in an interview with EVMS for its continuing education program here.
Collaborators:
Beth Cardier, Sirius-Beta Corp, PI
Niccolo Casas, RISD Rhode Island School of Design; University College of London
Ted Goranson, Sirius-Beta Corp
Alessio Erioli, Università di Bologna
Patric Lundberg, Richard Ciavarra and Larry Sanford, Eastern Virginia Medical School
Towards the ‘InnerNet’:
An integrated sensor analysis of biome/microbiome systems, employing novel interactivity through acoustics and design for personalized health monitoring.
This project is supported by a $100K seed grant.
The InnerNet project considers the whole biophysical system of the body with the goal of understanding how bodily systems ‘talk’ to one another by tapping into communications between the body and the microbiome. This team will explore the development of wearable external and internal sensor arrays for this purpose.
Collaborators:
Paul Weiss, University of California, Los Angeles, PI
Ruth West, University of North Texas
Andrea Polli, University of New Mexico
Beth Cardier, Sirius-Beta Corp
Niccolo Cassas, RISD Rhode Island School of Design; University College of London
Allison Kudla, Institute for Systems Biology